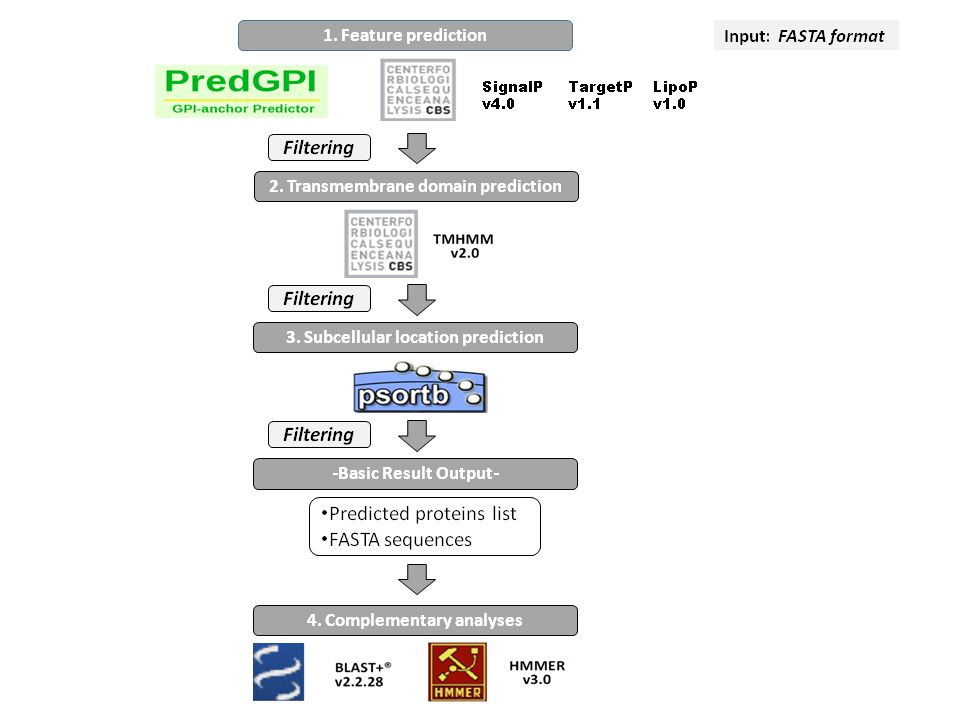
Prokaryotic secretion pipeline
This pipeline includes different tools for secretome analysis and a set of Perl/Bioperl and R scripts has been set up. Prokaryotic analysis is presented under a one-step query interface, where an input FASTA file should be uploaded. User can select the cut-off scores for the processing tools (or default values). Optional tests are chosen and input file is submited.
* (G.O. enrichment test is not available for this analysis)
* Mandatory information